The Physcraper results¶
Files generated by a Physcraper run¶
Within the output directory defined by the user, a Physcraper run generates various subdirectories which are labeled with a TAG name that corresponds to the file name of the input alignment.
The subdirectories consist of:
- Input files
Within the inputs_TAG directory, Physcraper writes tree and alignment files used in a Physcraper run for the sake of reproducibility, taxon name matching, and taxon reconciliation.
It also writes the “.config” file down if none was provided, as well as the results of the mapping of the tree taxon labels, saved as “otu_info.csv”
- Run files
Within the run_TAG directory, all run files are also automatically written down: Blast runs, alignments, RAxML trees, bootstrap results, etc.
Intermediate processing files, and the json formatted otu information are also stored here. Many fo these files are re-used in the event that the analysis crashes and is restarted. Make sure you use a new output directory or otherwise empty this folder if you want to modify the initial run parameters.
The trees are reconstructed using RAxML, with tip labels corresponding to local ids (e.g., otu42009, otuPS1) and not taxon names (e.g., Ceiba), nor taxonomic ids (e.g., ott or ncbi). Branch lengths are proportional to relative substitution rates.
- BLAST files
The blast_TAG folder contains XML files with BLAST results for each sequence in the input alignment.
- Output files
In the output_TAG folder, the updated tree with taxon names as tip labels is saved as “updated_taxonname.tre”.
A version of the updated tree in nexson format containing several types of tip labels is also saved here. From the nexson tree, a tree with any
label can be produced.
See section relabeling the trees below for more info on how to generate trees with different types of labels.
The “seqlen_mismatch.txt” file contains the acession numbers, taxa, and sequence lengths of BLAST matches that didn’t meet the sequence length cutoffs.
Finally, the CSV file “otu_info_TAG.csv” contains a summary of information about original and newly added sequences.
Visualizing the Physcraper results¶
There are several tree visualization tools available. For reproducibility purposes, we will use some handy functions from the R language to see our results.
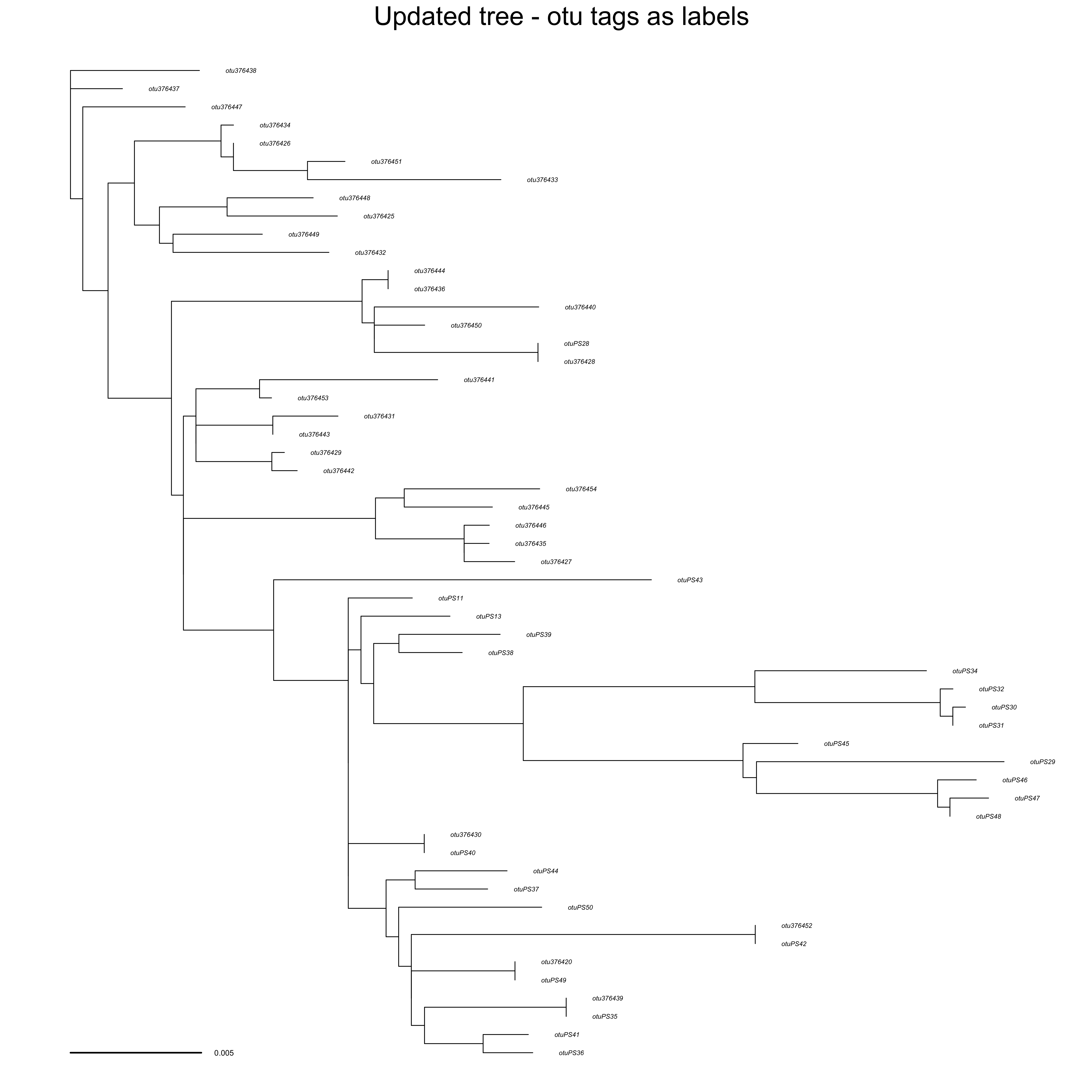
updated_tree_otus <- ape::read.tree(file = "docs/examples/pg_55/run_pg_55tree5864_ndhf/RAxML_bestTree.2020-06-18")
ape::plot.phylo(ape::ladderize(updated_tree_otus), type = "phylogram", cex = 0.25, label.offset = 0.001, edge.width = 0.5)
ape::add.scale.bar(cex = 0.3, font = 1, col = "black")
mtext("Updated tree - otu tags as labels", side = 3)
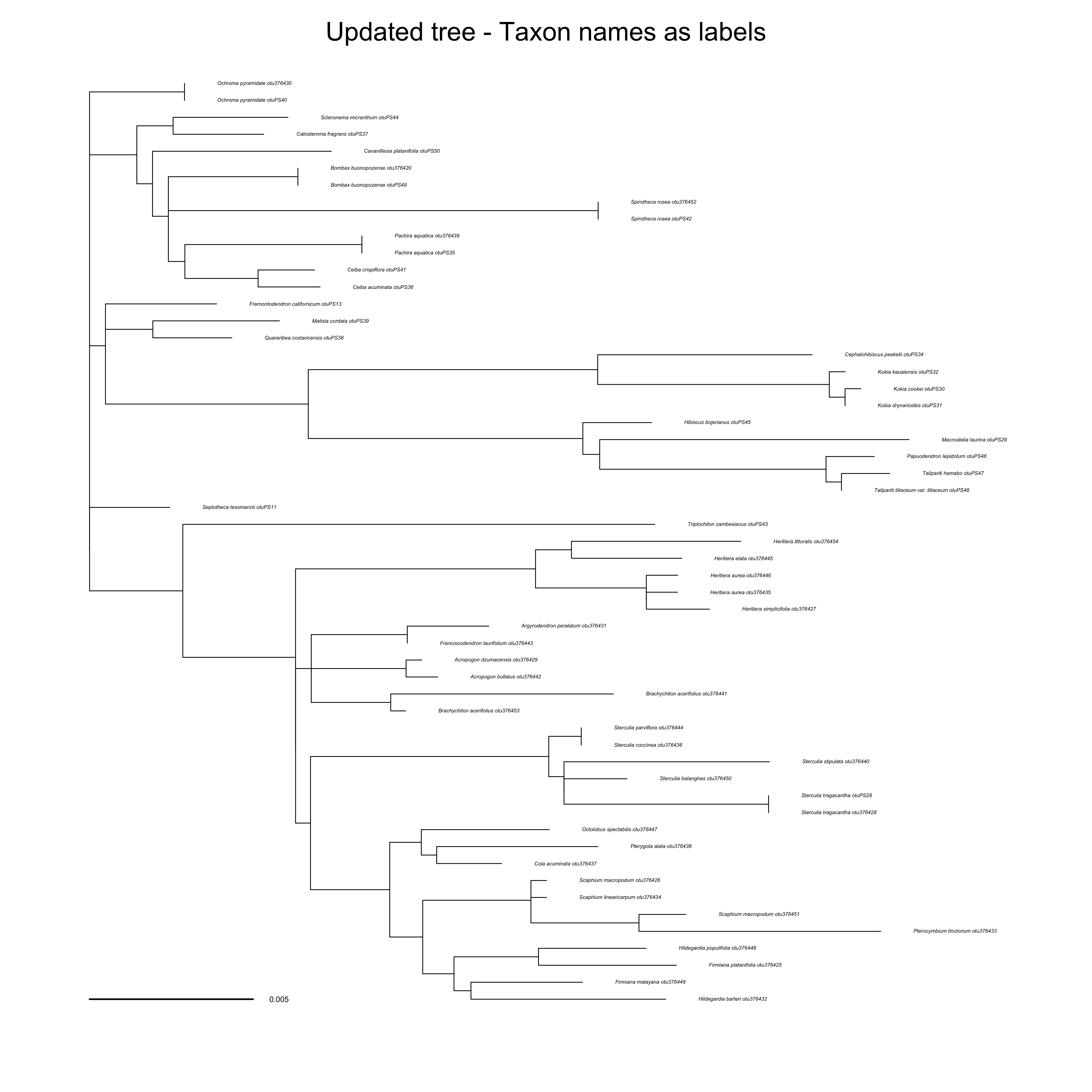
updated_tree_taxonname <- ape::read.tree(
file = "docs/examples/pg_55/outputs_pg_55tree5864_ndhf/updated_taxonname.tre")
ape::plot.phylo(ape::ladderize(updated_tree_taxonname), type = "phylogram", cex = 0.2, label.offset = 0.001, edge.width = 0.5)
ape::add.scale.bar(cex = 0.3, font = 1, col = "black")
mtext("Updated tree - Taxon names as labels", side = 3)
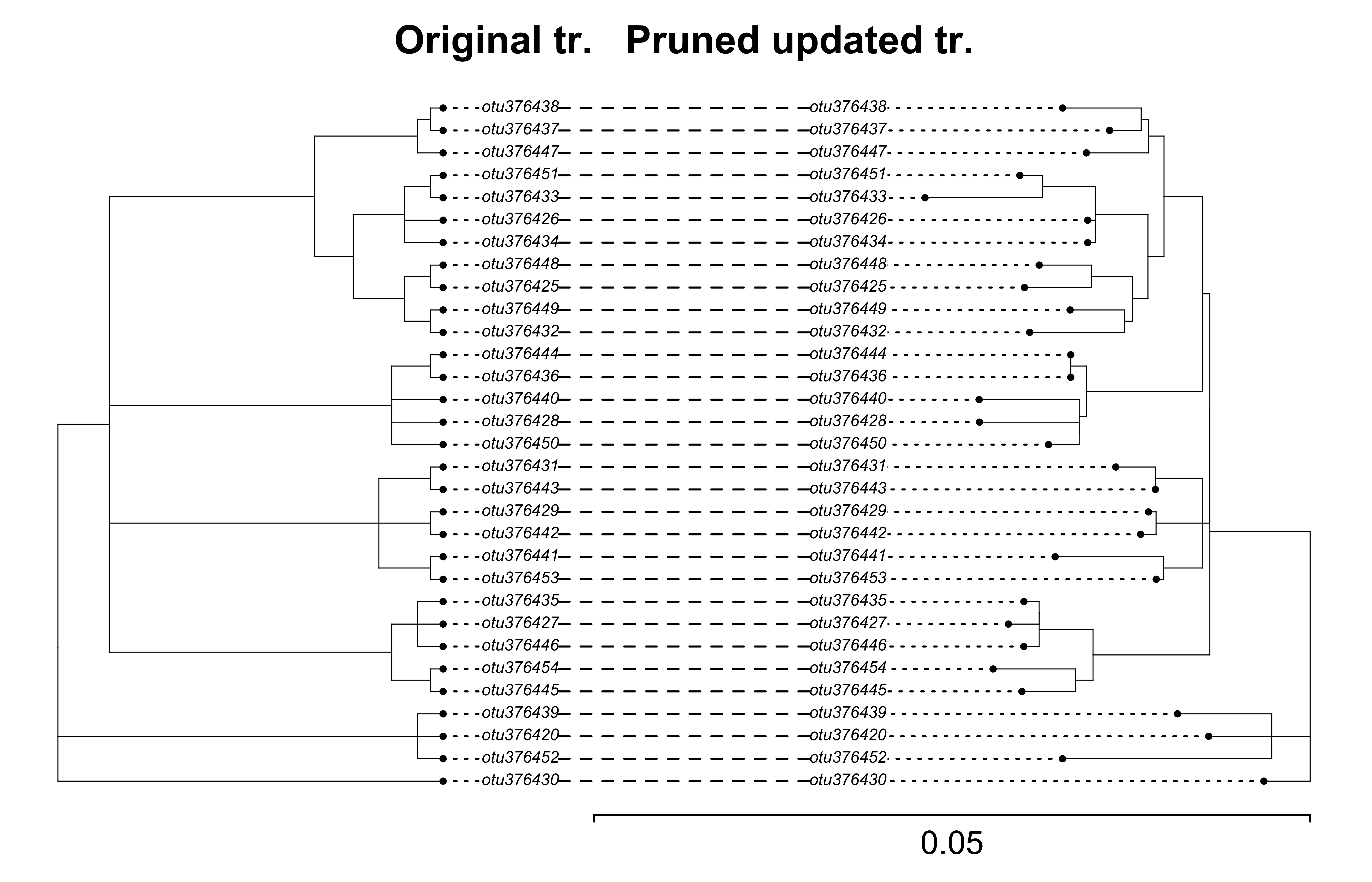
Compare the original tree with the pruned updated tree
original_tree_otus <- ape::read.tree(file = "docs/examples/pg_55/inputs_pg_55tree5864_ndhf/physcraper_pg_55tree5864_ndhf.tre")
updated_tree_otus_pruned <- ape::read.tree(
file = "../data/pg_55/pruned_updated.tre"
)
Now plot them face to face.
We can prune the updated tree, so it is a straight forward comparison:
cotree <- phytools::cophylo(original_tree_otus, updated_tree_otus_pruned, rotate.multi =TRUE)
Rotating nodes to optimize matching… Done.
phytools::plot.cophylo(x = cotree, fsize = 0.5, lwd = 0.5, mar=c(.1,.2,2,.3), ylim=c(-.1,1), scale.bar=c(0, .05))
title("Original tr. Pruned updated tr.", cex = 0.1)
But it is more interesting to plot it with all the new tips, so we see exactly where the new things are:
original_tree_taxonname <- ape::read.tree(file = "docs/examples/pg_55/inputs_pg_55tree5864_ndhf/taxonname.tre")
cotree2 <- phytools::cophylo(
original_tree_taxonname,
updated_tree_taxonname,
rotate.multi =TRUE)
Rotating nodes to optimize matching… Done.
phytools::plot.cophylo(
x = cotree2,
fsize = 0.3,
lwd = 0.4,
mar=c(.1,.1,2,.5),
ylim=c(-.1,1),
scale.bar=c(0, .05),
# link.type="curved",
link.lwd=3,
link.lty="solid",
link.col=phytools::make.transparent("#8B008B",0.5))
title("Original tree Updated tree", cex = 0.1)

We can also plot the updated tree against the synthetic subtree of Malvaceae, to visualize how it updates our current knowledge of the phylogeentic relationships within the family.
However, we are having some trouble with matching the tips right now! A fix will come soon:
tolsubtree <- rotl::tol_subtree(ott_id = 279960)
ape::Ntip(tolsubtree)
#> [1] 5898
grep("Pterygota_alata", tolsubtree$tip.label)
#> [1] 5714
updated_tree_taxonname$tip.label
#> [1] "Fremontodendron_californicum_otuPS13"
#> [2] "Quararibea_costaricensis_otuPS38"
#> [3] "Matisia_cordata_otuPS39"
#> [4] "Hibiscus_bojerianus_otuPS45"
#> [5] "Macrostelia_laurina_otuPS29"
#> [6] "Talipariti_tiliaceum_var._tiliaceum_otuPS48"
#> [7] "Talipariti_hamabo_otuPS47"
#> [8] "Papuodendron_lepidotum_otuPS46"
#> [9] "Cephalohibiscus_peekelii_otuPS34"
#> [10] "Kokia_kauaiensis_otuPS32"
#> [11] "Kokia_drynarioides_otuPS31"
#> [12] "Kokia_cookei_otuPS30"
#> [13] "Ochroma_pyramidale_otuPS40"
#> [14] "Ochroma_pyramidale_otu376430"
#> [15] "Catostemma_fragrans_otuPS37"
#> [16] "Scleronema_micranthum_otuPS44"
#> [17] "Cavanillesia_platanifolia_otuPS50"
#> [18] "Spirotheca_rosea_otuPS42"
#> [19] "Spirotheca_rosea_otu376452"
#> [20] "Bombax_buonopozense_otuPS49"
#> [21] "Bombax_buonopozense_otu376420"
#> [22] "Ceiba_acuminata_otuPS36"
#> [23] "Ceiba_crispiflora_otuPS41"
#> [24] "Pachira_aquatica_otuPS35"
#> [25] "Pachira_aquatica_otu376439"
#> [26] "Septotheca_tessmannii_otuPS11"
#> [27] "Triplochiton_zambesiacus_otuPS43"
#> [28] "Heritiera_elata_otu376445"
#> [29] "Heritiera_littoralis_otu376454"
#> [30] "Heritiera_aurea_otu376446"
#> [31] "Heritiera_simplicifolia_otu376427"
#> [32] "Heritiera_aurea_otu376435"
#> [33] "Brachychiton_acerifolius_otu376453"
#> [34] "Brachychiton_acerifolius_otu376441"
#> [35] "Acropogon_bullatus_otu376442"
#> [36] "Acropogon_dzumacensis_otu376429"
#> [37] "Franciscodendron_laurifolium_otu376443"
#> [38] "Argyrodendron_peralatum_otu376431"
#> [39] "Sterculia_balanghas_otu376450"
#> [40] "Sterculia_tragacantha_otu376428"
#> [41] "Sterculia_tragacantha_otuPS28"
#> [42] "Sterculia_stipulata_otu376440"
#> [43] "Sterculia_coccinea_otu376436"
#> [44] "Sterculia_parviflora_otu376444"
#> [45] "Hildegardia_barteri_otu376432"
#> [46] "Firmiana_malayana_otu376449"
#> [47] "Firmiana_platanifolia_otu376425"
#> [48] "Hildegardia_populifolia_otu376448"
#> [49] "Scaphium_linearicarpum_otu376434"
#> [50] "Scaphium_macropodum_otu376426"
#> [51] "Pterocymbium_tinctorium_otu376433"
#> [52] "Scaphium_macropodum_otu376451"
#> [53] "Octolobus_spectabilis_otu376447"
#> [54] "Cola_acuminata_otu376437"
#> [55] "Pterygota_alata_otu376438"
Trick for the cophylo titles and margins from https://cran.r-project.org/web/packages/phangorn/vignettes/IntertwiningTreesAndNetworks.html
Analysing the Physcraper results¶
Under construction
The functionalities described in the following sections are still under development. Even in this beta form, we think they have the potential to be useful, so we decided to document them here.
The first two functionalities are not accessible through the command line yet. This means that you can access them only through Python for the moment. The last functionality can be accessed through the command line, but it has not been fully tested yet. Use with caution.
Relabeling the trees¶
For downstream analyses and figure making, it can be handy to swap labels on tips of the updated phylogeny from alternative taxonomies or taxon id numbers.
Do that with the write_labelled function. For example, to change e.g.,
from physcraper import treetaxon
pg55 = treetaxon.generate_TreeTax_from_run('docs/examples/pg_55_web')
pg55.write_labelled(label='^ot:ottTaxonName', norepeats=False, path='tests/tmp/pg_55_repeats.tre')
Rerooting the trees¶
A correctly rooted phylogeny is needed to compare relationships between two or more phylogenetic hypotheses.
Automatic rooting of phylogenies is not straightforward. Physcraper’s root_tree_from_synth function places a suggested root based on relationships in OpenTree’s synthetic tree or in its taxonomic tree.
To root a Physcraper tree using either the OpenTree taxonomy, or the OpenTree synthetic tree. First load the tree object:
from physcraper import treetaxon
pg55 = treetaxon.generate_TreeTax_from_run('docs/examples/pg_55_web')
Then, to root based on the OpenTree taxonomy, set base = "ott":
from physcraper import opentree_helpers
ott_rooted_tree = opentree_helpers.root_tree_from_synth(pg55.tre, pg55.otu_dict, base='ott')
pg55.tre = ott_rooted_tree # set tree rooted based on taxonomy as the tree object
pg55.write_labelled(label="^ot:ottTaxonName", path="tests/tmp/pg_55_ott_root.tre")
And, to root based on phylogenetic relationships in the OpenTree synthetic tree, set base = "synth":
from physcraper import opentree_helpers
synth_rooted_tree = opentree_helpers.root_tree_from_synth(pg55.tre, pg55.otu_dict, base='synth')
pg55.tre = synth_rooted_tree # set tree rooted based on synth as the tree object
pg55.write_labelled(label="^ot:ottTaxonName", path="tests/tmp/pg_55_synth_root.tre")
In this example both trees are the same even though they use the MRCA of different pairs of taxa, because those MRCA’s map to the same node on the output tree.
However rooting based on OTT can be unreliable, especially if taxonomy is a poor fit to true evolutionary relationships.
So whenever possible, the root should be specified by the user, for example by choosing from tips in the “otu_info.csv” file in the outputs folder of a Physcraper run, e.g.,
pg55 = treetaxon.generate_TreeTax_from_run('docs/examples/pg_55_web')
outgroup = ['otu376436','otu376444']
mrca = pg55.tre.mrca(taxon_labels=outgroup)
pg55.tre.reroot_at_node(mrca, update_bipartitions=True)
pg55.write_labelled(label="^ot:ottTaxonName", path="tests/tmp/pg_55_manual_root.tre")
Tree comparison with Robinson-Foulds (RF) distance¶
The tree_comparison.py script takes as an argument the output directory of a Physcraper run,
and compares the relationships in the final tree to the relationships in the input tree.
Usage:
tree_comparison.py [-h] [-d DIRECTORY_NAME] [-t1 FILE_NAME] [-t2 FILE_NAME] [-otu FILE_NAME] [-og OUTGROUP] [-o DIRECTORY_NAME]
Arguments:
- -h , --help
Show the help message and exit.
- -d DIRECTORY_NAME, --dir DIRECTORY_NAME
A name (and path) to a Physcraper output directory.
- -o DIRECTORY_NAME, --output DIRECTORY_NAME
Name (and path) for a directory to write the results of the comparison analysis to. If it exists, it will be overwritten.
- -otu FILE_NAME, --otu_info FILE_NAME
Name (and path) of JSON file containing taxon information. Stored in the Physcraper output directory
runfolder.- -t1 FILE_NAME, --tree1 FILE_NAME
File name (and path) of original tree from a Physcraper output directory
inputsfolder. Alternatively, any file containing a tree to compare to -t2.- -t2 FILE_NAME, --tree2 FILE_NAME
File name (and path) of an updated tree from a Physcraper output directory
outputsfolder. Alternatively, any file containing a tree to compare to -t1.
This is the simplest command line for comparison of two trees:
tree_comparison.py [-h] [-d DIRECTORY_NAME] [-o DIRECTORY_NAME]
For example:
tree_comparison.py -d docs/examples/pg_55_web/ -o pg_55_comparison
It compares the original tree from the inputs folder and the updated tree from the outputs folder.
It uses the rooting functions described above to ensure the two trees have the same root.
By default, it will root trees based on the OpenTree Taxonomy.
Alternatively, you can pass in OpenTree taxonomic ids (OTT ids) of two or more taxa from the input tree to use as outgroups to root both trees.
For example:
tree_comparison.py -d docs/examples/pg_55_web/ -og otu376420 otu376439 otu376452 -o pg_55_comparison
If the comparison between the two trees is possible (outgroup-wise), the script will print the results to screen, including:
- The number of new tips
- The number of new taxa
- Whether the taxa in the tree are included synthesis phylogenies currently in OpenTree
- Which taxa phylogenetic information is not currently incorporated into the synthetic tree
- The RF distance and weighted RF distance between the relationships of tips that are in both trees
- How the estimates of phylogenetic relationships of taxa included in the OpenTree taxonomy from both trees have conflict with the monophyly of the OpenTree synthetic tree.